pdb2blend conversion toolby Malte Reimold 2006Version 1.2 is released!
Pdb2blend is a python script for the open source 3d modeling software blender. examples
features
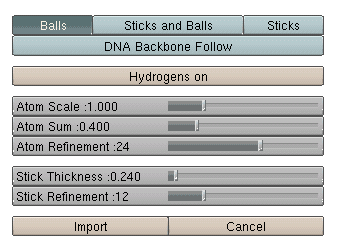
modes:1. Ballscreates a ball model with overlapping spheres when using the standard parameters the ball sizes can be adjusted by choosing a constant scaling factor (atom scale) as well as a constant summand (atom sum) that is added to the ball size. With atom scale = 1 and atom sum = 0 one will get the covalent radius for any atom defined. With atom scale = 0 and atom sum > 0 the model will have uniform ball sizes. 2. Stickscreates a stick model. The stick diameter can be chosen here (Stick Thickness). Until now the regular stick mode (as well as stick and ball) does only work for less than 10000 atoms. 3. Sticks and BallsA combination of ball and stick mode. When using standard parameters the atoms are scaled down a little bit. 4. DNA Backbone FollowWhen turning on this option a stick model will be formed connecting the phosphorous atoms of oligonucleotide backbones. This works even for systems with more than 10000 atoms. Model refinement:The refinement of spheres and sticks can be chosen. With higher values the model becomes more detailed. filesNew Version pdb2blend v1.2 is now available!
Download pdb2blend12 as zip file. |
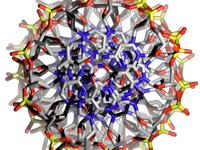
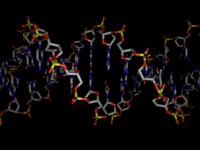

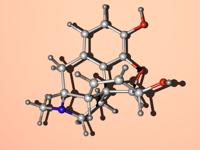

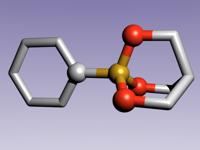
 malte.reimold.de.
malte.reimold.de.